The long-standing practice of data sharing in genomics can be traced to the Bermuda principles, which were formulated during the human genome project (Contreras, 2010). While the Bermuda principles focused on open sharing of DNA sequence data, they heralded the adoption of other open source standards in the genomics community. For example, unlike many other scientific disciplines, most genomics software is open source and this has been the case for a long time (Stajich and Lapp, 2006). The open principles of genomics have arguably greatly accelerated progress and facilitated discovery.
While open sourcing has become de rigueur in genomics dry labs, wet labs remain beholden to commercial instrument providers that rarely open source hardware or software, and impose draconian restrictions on instrument use and modification. With a view towards joining others who are working to change this state of affairs, we’ve posted a new preprint in which we describe an open source syringe pump and microscope system called poseidon:
A. Sina Booeshaghi, Eduardo da Veiga Beltrame, Dylan Bannon, Jase Gehring and Lior Pachter, Design principles for open source bioinstrumentation: the poseidon syringe pump system as an example, 2019.
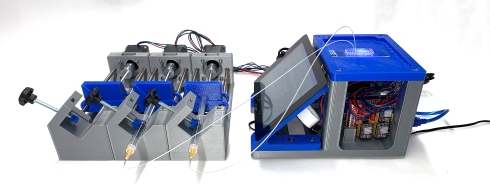
The poseidon system consists of
- A syringe pump that can operate at a wide range of flow rates. The bulk cost per pump is $37.91. A system of three pumps that can be used for droplet based single-cell RNA-seq experiments can be assembled for $174.87
- A microscope system that can be used to evaluate the quality of emulsions produced using the syringe pumps. The cost is $211.69.
- Open source software that can be used to operate four pumps simultaneously, either via a Raspberry Pi (that is part of the microscope system) or directly via a laptop/desktop.

Together, these components can be used to build a Drop-seq rig for under $400, or they can be used piecemeal for a wide variety of tasks. Along with describing benchmarks of poseidon, the preprint presents design guidelines that we hope can accelerate both development and adoption of open source bioinstruments. These were developed while working on the project; some were borrowed from our experience with bioinformatics software, while others emerged as we worked out kinks during development. As is the case with software, open source is not, in and of itself, enough to make an application usable. We had to optimize many steps during the development of poseidon, and in the preprint we illustrate the design principles we converged on with specific examples from poseidon.
The complete hardware/software package consists of the following components:
- Hardware
- STL files (needed for 3D printing) and CAD design files. STL files are also available via Thingiverse where there are links to 3rd party vendors that can print and ship the designs. The CAD files can be edited and modified.
- CAD visualizations for the pump and the microscope. These are useful when building the hardware as they show what the completed products look like.
- Bill of materials (all parts can be sourced from Amazon and links are provided). Pricing is broken down by unit cost, system cost, and bulk cost.
- Assembly videos for every component of the system.
- Software
- Open source software available on GitHub.
- Startup checklist.
- Getting started video showing how to run the system for the first time.
We benchmarked the system thoroughly and it has similar performance to a commercial Harvard Apparatus syringe pump; see panel (a) below. The software driving the pumps can be used for infusion or withdrawl, and is easily customizable. In fact, the ability to easily program arbitrary schedules and flow rates without depending on vendors who frequently charge money and require firmware upgrades for basic tasks, was a major motivation for undertaking the project. The microscope is basic but usable for setting up emulsions. Shown in panel (b) below is a microfluidic droplet generation chip imaged with the microscope. Panel (c) shows that we have no trouble generating uniform emulsions with it.

Together, the system constitutes a Drop-seq rig (3 pumps + microscope) that can be built for under $400:
We did not start the poseidon project from scratch. First of all, we were fortunate to have some experience with 3D printing. Shortly after I started setting up a wet lab, Shannon Hateley, a former student in the lab, encouraged me to buy a 3D printer to reduce costs for basic lab supplies. The original MakerGear M2 we purchased has served us well saving us enormous amounts of money just as Shannon predicted, and in fact we now have added a Prusa printer:

The printer Shannon introduced to the lab came in handy when, some time later, after starting to perform Drop-seq in the lab, Jase Gehring became frustrated with the rigidity commercial syringe pumps he was using. With a 3D printer available in-house, he was able to print and assemble a published open source syringe pump design. What started as a hobby project became more serious when two students joined the lab: Sina Booeshaghi, a mechanical engineer, and Eduardo Beltrame, an expert in 3D printing. We were also encouraged by the publication of a complete Drop-seq do-it-yourself design from the Satija lab. Starting with the microscope device from the Stephenson et al. paper, and the syringe pump from the Wijnen et al. paper, we worked our way through numerous hardware design optimizations and software prototypes. The photo below shows the published work we started with at the bottom, the final designs we ended up with at the top, and intermediate versions as we converged on design choices:

In the course of design we realized that despite a lot of experience developing open source software in the lab, there were new lessons we were learning regarding open-source hardware development, and hardware-software integration. We ended up formulating six design principles that we explain in detail in the preprint via example of how they pertained to the poseidon design:

We are hopeful that these principles we adhered to will serve as useful guidelines for others interested in undertaking open source bioinstrumentation projects.

16 comments
Comments feed for this article
January 19, 2019 at 2:34 am
Francisco Lima
Nice story. There was a nice feature in a 2018 Nature issue covering the topic. I look forward to see more about the project.
January 20, 2019 at 6:09 pm
Viraj Shah
This is awesome. Thank you for releasing this. Had I discovered this a day sooner, I would have stopped working through another open source design : https://chem.uncg.edu/croatt/flow-chemistry/
While the above link is well documented, it misses a few things like what kinds of syringes and motor controller boards were used. I ended up spending a few hours on Pololu looking at stepper motor drivers and then I saw this project on Hacker News. This is phenomenally well documented for an open source project and I love that you did the comparison with a commercial tool. I look forward to building these pumps for my own microfluidic devices. Thanks again and hats off to you and the team.
January 20, 2019 at 9:23 pm
Yosi
If I’m not wrong, this is ~400$ not including the 3D printer. Also, it does not including the time/effort/cost it takes to master the 3D printer.
How much would you evaluate these extra steps?
January 20, 2019 at 10:20 pm
Lior Pachter
Owning and operating a 3D printer is not necessary to obtain the printed design. One can print the poseidon system directly from the Thingiverse page with a few different vendors with prices ranging from $30-$70: https://www.thingiverse.com/thing:3074853/apps/print/#apps
Alternatively one can upload the STL files to one of many other websites that will ship the printed design for a fee. Furthermore, 3D printers are available for free printing in many public / university libraries, where there are also courses/staff available to assist in printing.
January 21, 2019 at 1:00 am
Viraj Shah
On a slightly related note, I might have missed it but do you have information on the material (e.g. PLA, ABS, or other) used for 3D printing? And at what infill percentage you printed the parts?
I only ask because due to the potentially high fluidic resistances in microfluidic devices, I want to ensure that I print a stiff enough structure to prevent any bending and thus inaccurate flow rates. Thanks.
January 21, 2019 at 12:00 pm
Eduardo Da Veiga Beltrame
To answer to Viraj Shah’s questions below:
All the prints we have done were using PLA 40-60% infill, layer heights of 0.2mm, 0.25mm and 0.3mm.
I would suggest printing at 50% or higher infill. If you want to print with another material like ABS or PET to make the pumps slightly stronger, I see no problem with that, but we have not tried it and validated if there are any issues with the design tolerances (worst case scenario you’ll need to widen the screw holes a little bit after the print). To make sure that the carriage won’t bend under high syringe pressures, ensure that the linear rods are securely attached to the printed body (I have previously super-glued them in place and that works, but is a bit janky, which is why we came up with the pressing screw design)
If you are looking into working with high pressures, I think that your main concern will be making sure the stepper motor won’t stall. My suggestion in that case is to use a geared high torque stepper motor( for example here’s a 100:1 one: https://www.amazon.com/100-Planetary-Gearbox-Stepper-Torque/dp/B00QEWAL98/ref=sr_1_1?keywords=geared+stepper+motor&qid=1548096784&sr=8-1)
They are a bit more expensive (~$50 each) but are very very strong and will almost certainly suffice for your application (we tinkered with some of them before realizing 1/32 microstepping would do the trick with normal steppers). The one drawback is that the maximum speed will be limited so the motor may take a little bit to reach the desired positions when you are loading the syringes (unfortunately we couldn’t devise a simple and robust way to disengage the sled for loading the syringe, so you must move it to position).
If you have any other questions let us know, seeing the great response to the pumps I’m collecting all these questions to maybe make a FAQ on the poseidon website.
January 21, 2019 at 12:23 pm
Viraj Shah
Thanks Eduardo, this is very helpful! Awesome stuff. Will send any feedback I have via comments or email.
January 21, 2019 at 8:24 am
HardwareX (@HardwareX1)
Very nice work – many other open hardware systems being developed that may be of interest to bio scientists available http://www.appropedia.org/Open-source_Lab#Examples
April 8, 2019 at 5:09 pm
Artem Barski
This is really cool!
I was wondering if you guys tried to do Drop-seq or InDrop with this setup? Do you need a magnetic mixer to agitate cells/beads?
May 10, 2019 at 5:03 pm
Eduardo Da Veiga Beltrame
Hi Artem, we have not performed drop-seq with the poseidon system, but you would need magnetic mixer next to the syringes because the drop-seq beads are made of polystyrene, so they are dense and will quickly settle. The minidrops developed by the Satija group (https://www.nature.com/articles/s41467-017-02659-x) does have a magnetic stirrer for that purpose.
The inDrops beads are made of hydrogen, so they are not as dense and mixing is not critical. We’ve used the system to generate emulsions with inDrops droplet generating chips.
May 17, 2019 at 3:27 pm
Artem Barski
Thanks a lot, Eduardo! And the cells do not need a mixer too? Can you guys please post a photo of your inDrop set up? Also where do you get chips and beads? 1CellBio?
May 16, 2019 at 8:02 am
Uri David Akavia
One question I have on this – how do you make the microfluidic chips?
I understand that in commercial systems, the cost of consumable chips can be really large, so setting up your own system and buying chips would not be that helpful. I might be wrong about this, but I’m curious.
November 1, 2019 at 8:55 am
Marek Mutwil
Hi Lior and Eduardo,
Great work!
I am super hyped to get Drop-seq and poseidon established in my lab and I look forward to assembling it when the parts arrive next week.
Can you recommend a chip provider (Nanoshift, FlowJEM, uFluidix or other?)? Is the rest of the protocol (syringes, tubing) the same as other open-source solutions?
Cheers,
Marek
February 13, 2021 at 2:56 pm
Anton
This is probably the dumbest project one could launch in modern biotech-related research. With optimized single-cell technologies available, and given you want to compete with the high-quality research that the big labs produce on a daily basis, there is no room for any custom approaches. Use what is commercially available, focus on the science that comes with the data, and do not waste time building this kind of custom apparatus to get your single-cell stuff running. I would never ever even consider to use anything outside of what is commercially available and therefore is well-tested plus comes with warranty. Completly useless project.
February 13, 2021 at 3:15 pm
Lior Pachter
Actually, I’m not particularly interested in competing with the “high-quality research that big labs produce on a daily basis.” In the words of David Blackwell, who described himself as a “dilettante”, I would say that “basically, I’m not interested in doing research and I never have been…I’m interested in understanding, which is quite a different thing.”
February 13, 2021 at 6:29 pm
gasstationwithoutpumps
Lots of people are building “picroscopes” (digital microscopes using a Raspberry Pi). I understand that the “braingeneers” project at UCSC has built a 36-well version for monitoring growth of organoids without having to take them out of an incubator.